GRIFFIN
implicit force grid
2021
René Staritzbichler
GRIFFIN
Molecular dynamics
- point like atoms
- Newtons equation of motion
- highly accurate, yet very time-consuming
- small timesteps
- large number of pair interactions
- cannot resolve clashes in starting conformation
Speed MD up
Overcoming time and size limitations by simplification
- molecule: coarse grained simulations
- environment: implicit waters
Implicit water
- no water molecules actually present during simulation
- instead describe statistical effect
- potential of mean force
- exposure
Griffin
Implicit (potential) protein force field
- precalculated
- forces caused by protein are stored on grid
- atomic detail
- very fast
- simplification: static protein
Potential forces
\[ \vec{F} = q_{dummy} \cdot \sum_{i=1}^{atoms} \frac{1}{4\pi \epsilon_0} \frac{q_i}{r_{i,dummy}^2} \frac{\vec{r}}{r} \]
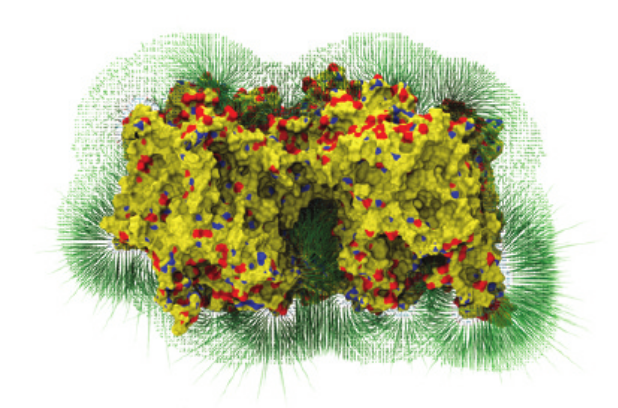
Force grid
\[ \vec{P} = \sum_{i=1}^{atoms} \frac{1}{4\pi \epsilon_0} \frac{q_i}{r_{i,gridpoint}^2} \frac{\vec{r}}{r} \]
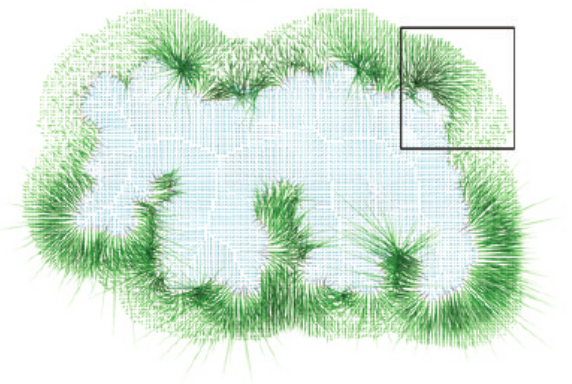
Forces
\[ \vec{P} = \sum_{i=1}^{atoms} \frac{1}{4\pi \epsilon_0} \frac{q_i}{r_{i,gridpoint}^2} \frac{\vec{r}}{r} \]
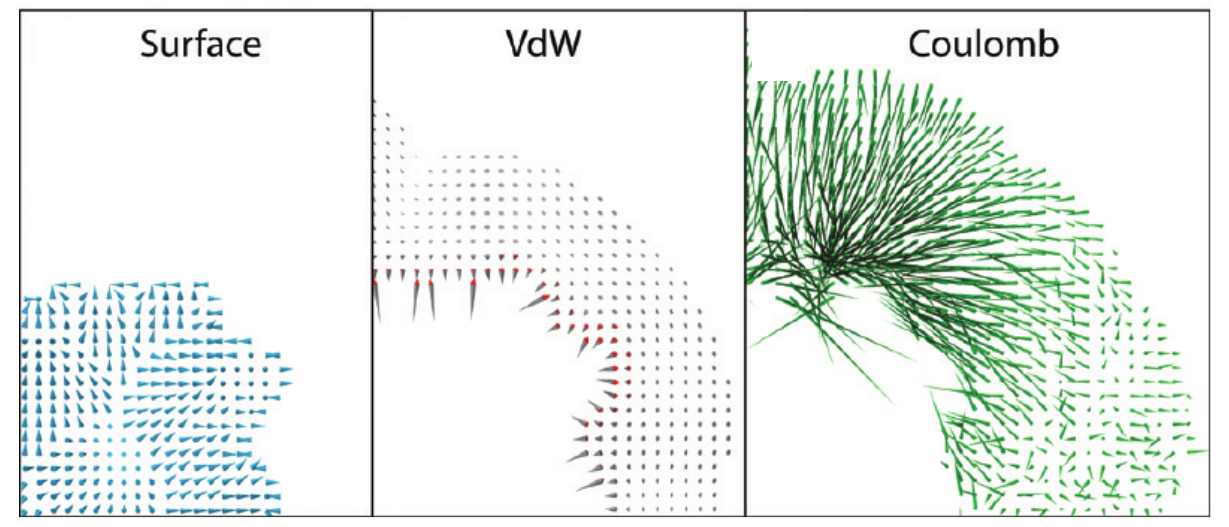
Resulting forces
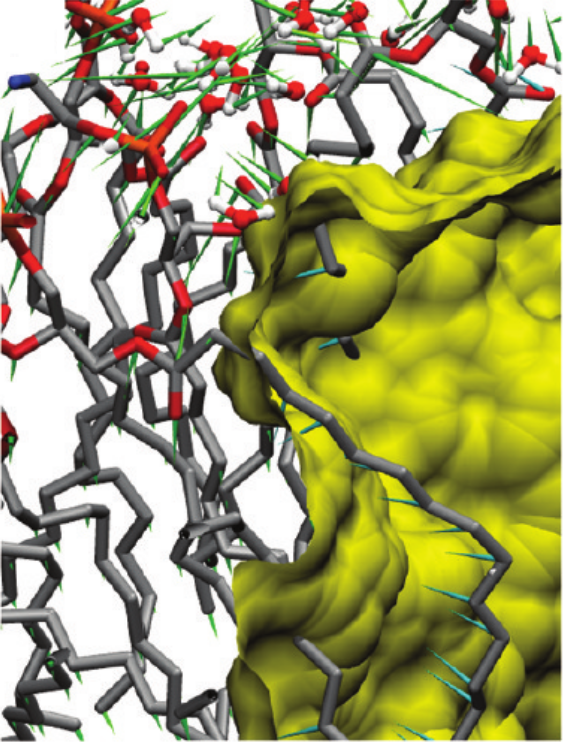
Running MD & Griffin
MD with a ghost force field
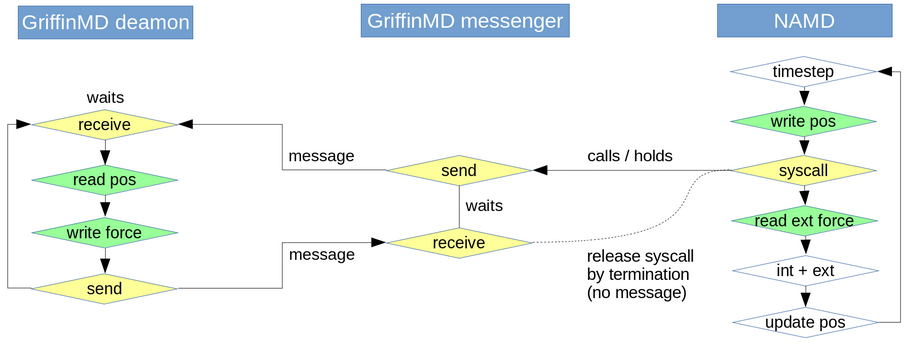
(-: In action :-)
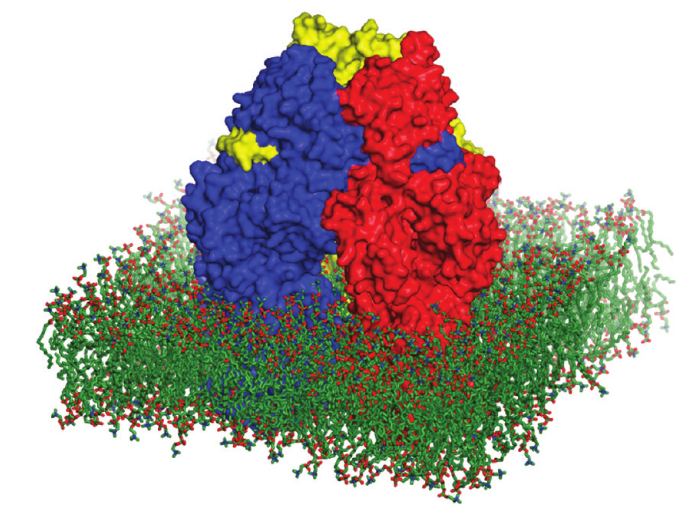

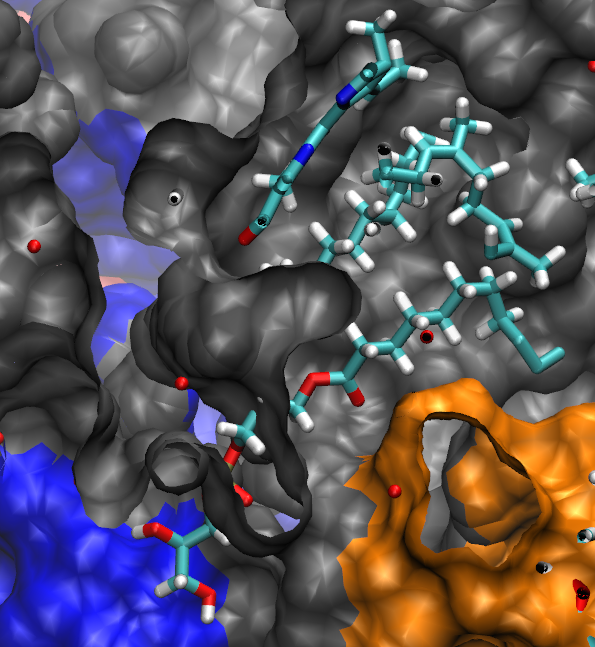
More action

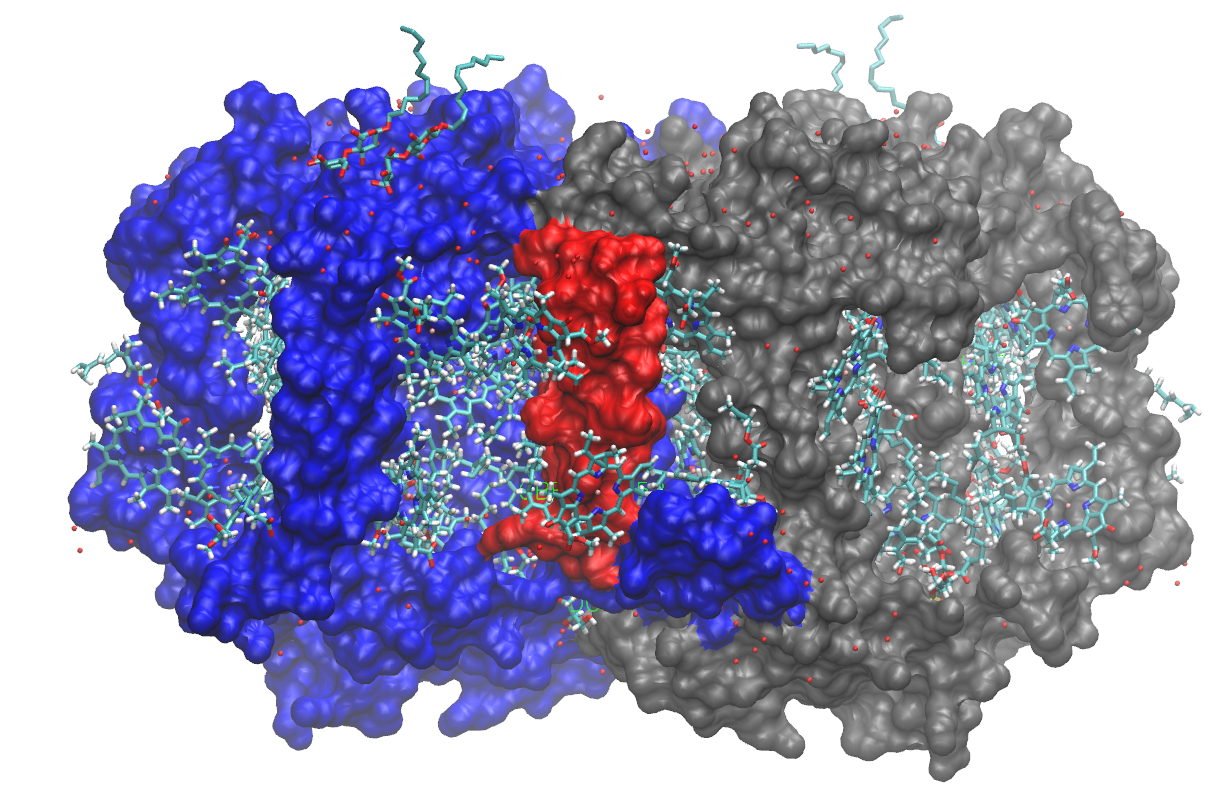
Photosystem